translation mode example¶
The BamQuery translation mode was designed to search BAM files from Ribo-seq data. In this mode, BamQuery can be used as a means to verify the presence of ribosome profiling reads that overlap with peptide MCSs.
Aware that the length of Ribo-seq reads varies between ~24-32 nucletotides, BamQuery counts reads according to their percentage overlapping with the MCS. For example, if a Ribo-seq read overlaps with 70% of the MCS in a given region, BamQuery counts this read as 0.7 instead of 1.
Note
BamQuery counts all reads for which the overlap with the MCSs is equal or higher to 60%.
Input¶
In this mode, instead of BAM_directories.tsv BamQuery expects a BAM_ribo_directories.tsv that includes the Ribo-seq datasets.
Command line:
BamQuery.py ./translation_example/Input translation_example v38_104 --mode translation
Input folder path_to_input_folder must containt the files : BAM_ribo_directories.tsv and peptides.tsv.
Output¶
BamQuery creates an output directory in the same path as the input folder.
This directory contains 3 folders and the main results are organized as follows:
├── alignments
│ ├── missed_peptides.info
│ └── translation_example_info_alignments.xlsx
├── logs
│ ├── BamQuery_Res_translation_example.log
│ └── Information_BAM_directories.log
├── plots
│ └── heat_maps
│ └── translation_evidence_heatmap
│ ├── average_translation_expression_heatmap
│ │ ├── norm_info.csv
│ │ ├── translation_example_ribo_norm_all_tissues.pdf
│ │ └── translation_example_ribo_norm_selected_tissues.pdf
│ └── total_translation_expression_heatmap
│ ├── translation_example_ribo_counts.csv
│ ├── translation_example_ribo_counts.pdf
│ ├── translation_example_ribo_norm.csv
│ └── translation_example_ribo_norm.pdf
└── res_translation
└── translation_example_ribo_count_info.xlsx
The output files (alignments and logs) in BamQuery translation mode are similar to those in normal mode, see `output_normal_mode_example`_ for detailed information about the output files : missed_peptides.info, sc_example_info_alignments.xlsx and logs.
plots¶
plots
└── heat_maps
└── translation_evidence_heatmap
├── average_translation_expression_heatmap
│ ├── norm_info.csv
│ ├── translation_example_ribo_norm_all_tissues.pdf
│ └── translation_example_ribo_norm_selected_tissues.pdf
└── total_translation_expression_heatmap
├── translation_example_ribo_counts.csv
├── translation_example_ribo_counts.pdf
├── translation_example_ribo_norm.csv
└── translation_example_ribo_norm.pdf
heat_maps¶
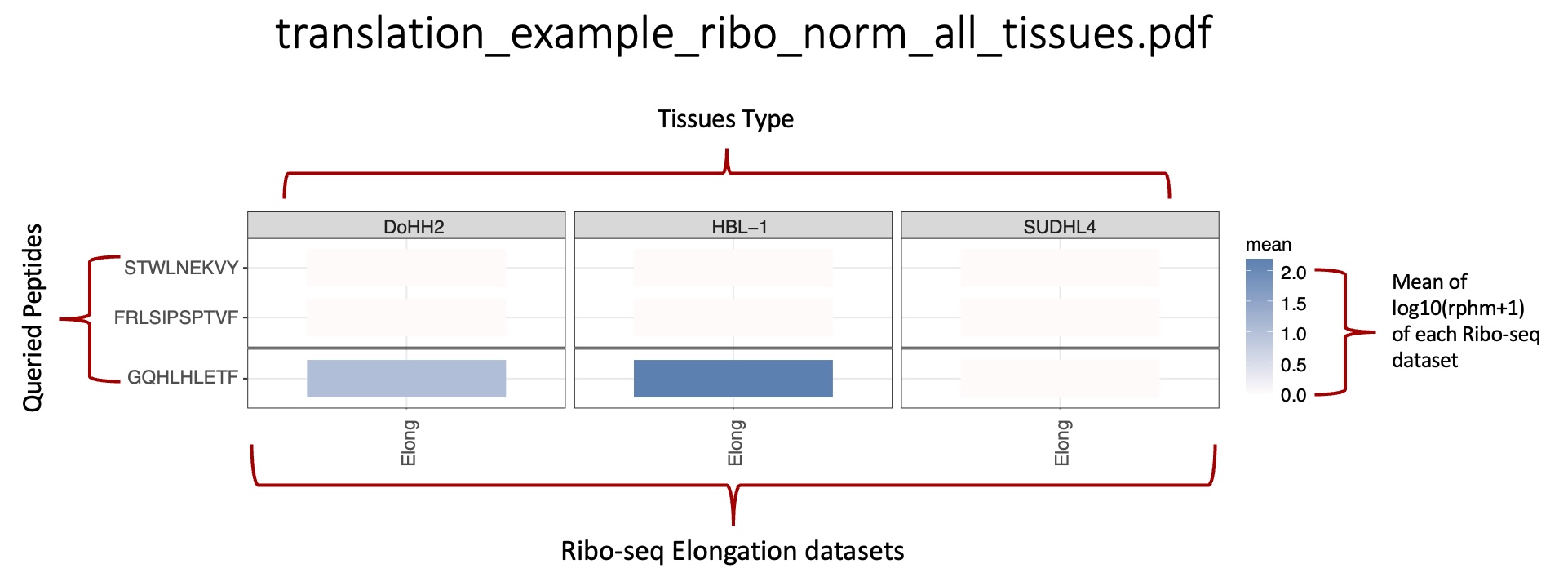
This folder contains the heat maps representing the translation expression level of all peptides queried.
average_translation_expression_heatmap folder: |br| Heat maps representing the mean translation expression for each peptide queried in the tissues associated with the BAM/CRAM ribo-seq files.
norm_info.csv: reports, for each peptide queried, the mean and median values of rphm in the tissues associated with the BAM/CRAM ribo-seq files.
_norm_all_tissues.pdf : Heat map representing the mean level of translation expression associated with tissue types, computed from all samples in the tissue.
_norm_selected_tissues.pdf: Heat map representing the mean level of translation expression associated with tissue types, computed from selected tissues (short list of tissues).
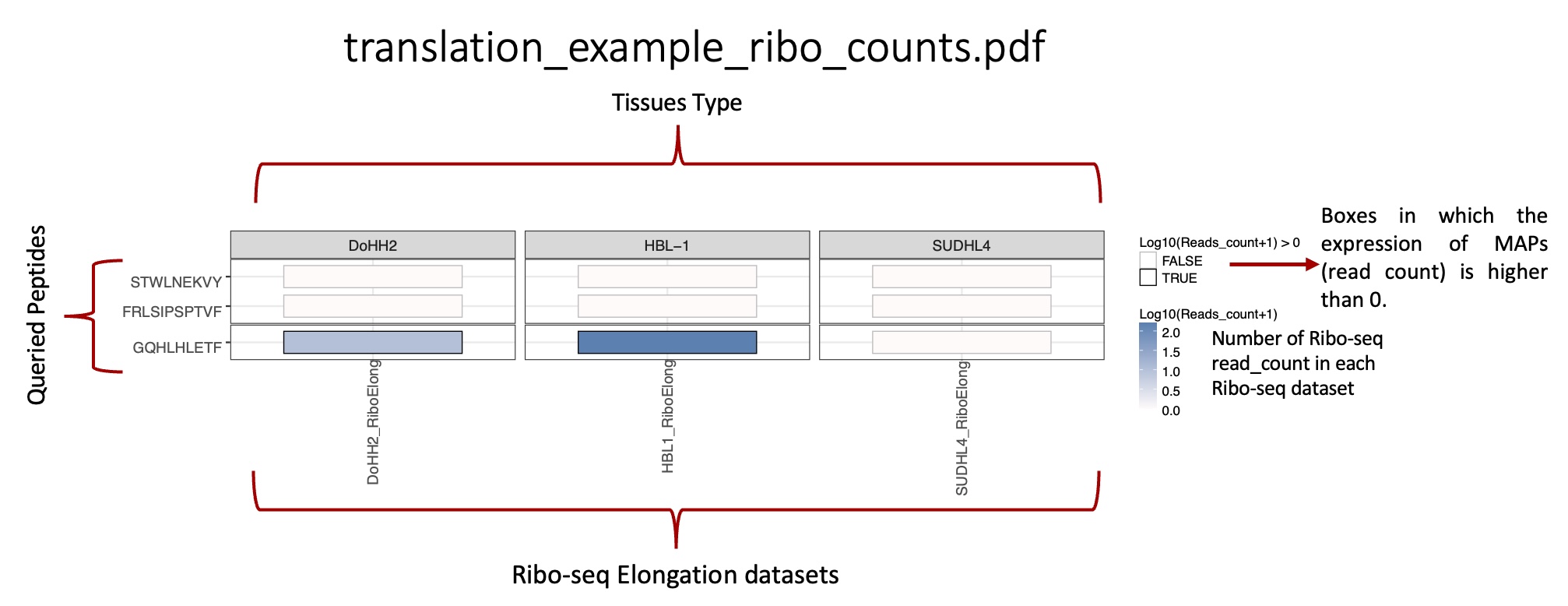
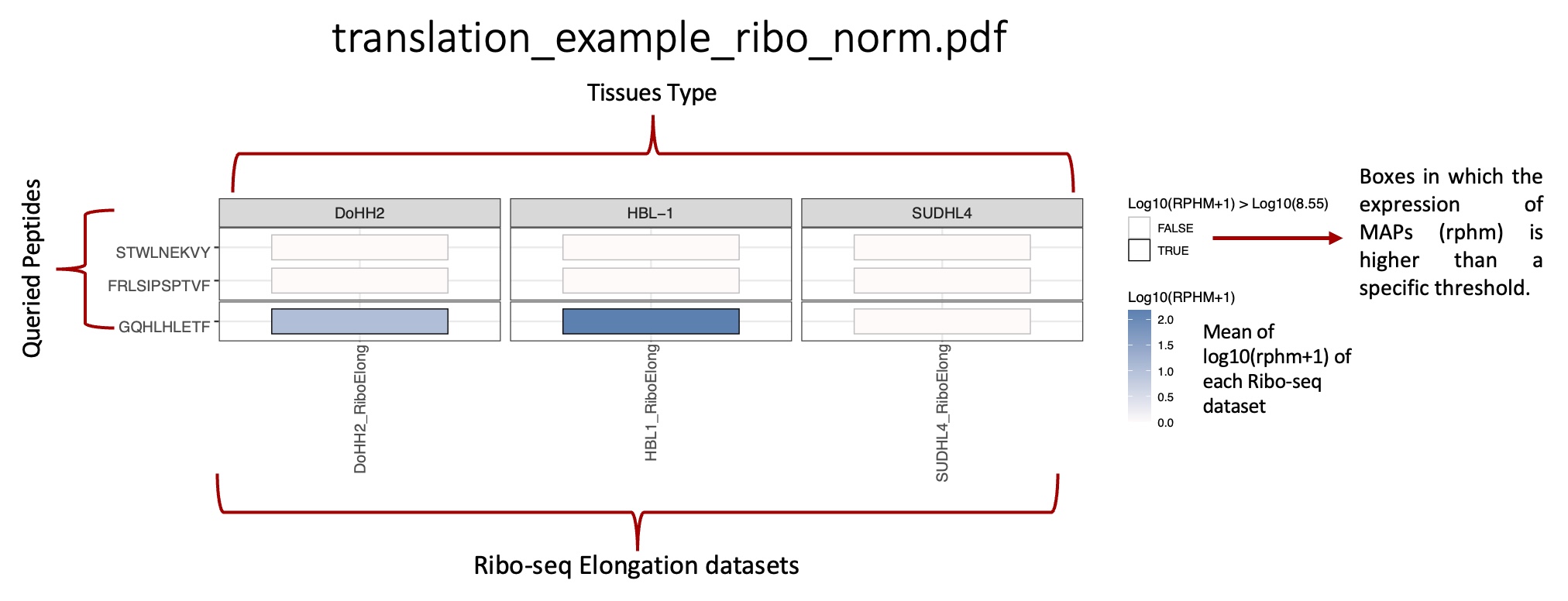
total_translation_expression_heatmap folder: |br| Heat maps representing the mean translation expression and total number of ribo-seq reads for each peptide queried in each BAM/CRAM ribo-seq file.
_ribo_counts.csv: reports, for each peptide queried, the total number of ribo-seq reads in each BAM/CRAM ribo-seq file.
_ribo_counts.pdf: heat map representing the mean number of ribo-seq reads in each BAM/CRAM ribo-seq file.
_ribo_norm.csv: reports, for each peptide queried, the mean rphm values in each BAM/CRAM ribo-seq file.
_ribo_norm.pdf: heat map representing the mean translation expression level of each peptide in each BAM/CRAM ribo-seq file.
Warning
Heat maps are produced for searches with less than 400 peptides.
Heat map in total_translation_expression_heatmap is produced only if the number of BAM/CRAM files queried are less than 100 tissues.